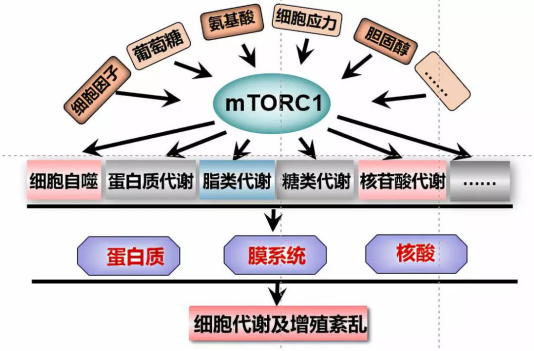
MTORC1, as a sensor of nutrition signal in
microenvironment, can sense amino acid, growth factor, glucose, cholesterol and
other signals in microenvironment, and then regulate several key processes in
cell and organism, such as glucose metabolism, protein metabolism, lipid
metabolism, cell autophagy and so on [1, 2].
However, when the key proteins of mTORC1 signaling pathway (mTOR, GATOR, PTEN,
TSC, LKB, AMPK, etc.) mutate, it will lead to the disorder of mTORC1 activity,
and then lead to the disorder of cell metabolism and cell proliferation, which
will also be the main inducement of many metabolic related diseases [3, 4]. It can be seen that mTORC1 signaling pathway
is the core of maintaining the metabolic balance of the body, and also an
important frontier field of cell metabolism. Further exploration of the
regulatory mechanism of mTORC1 signaling pathway can provide important theories
for molecular diagnosis, accurate classification, prognosis analysis and
targeted therapy based on metabolic diseases in clinic.
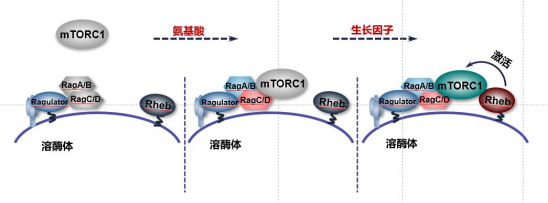
The activation of mTORC1 is mainly due to the
synergistic action of amino acid and growth factor signal, in which amino acid
signal mainly regulates the localization of mTORC1 on lysosomes through Rags
complex, while growth factor signal mainly promotes the activity of mTORC1 by
activating Rheb [2,5].
Prof. Wang Ping’s research group has been focusing on
the interaction mechanism between tumor cells and microenvironment, a key
scientific question in tumor biology for a long time, and discusses
ubiquitination regulation of tumor microenvironment systematically. A large
number of evidences have shown that ubiquitination plays an important role in
tumorigenesis. As an important post-translational modification of proteins,
ubiquitination (including sumoylation) has always been the focus and hotspot of
tumor biology research. Ubiquitination is a reversible enzyme cascade reaction,
which is precisely regulated by ubiquitin ligases and depubiquitinases. So far,
nearly 600 E3 ubiquitin ligases and 100 depubiquitinases have been reported[6-8]. However, the relationship between the most E3
ubiquitin ligases and depubiquitinases and tumorigenesis is not clear yet.
So, around the scientific question of "whether
ubiquitination participates in the nutrition signal in mTORC1 sensing
microenvironment", Wang Ping group of Tongji University School of Medicine
and Hui Kuanlin group of MD Anderson Cancer Center published papers "back
to back" in 2015, and found that ubiquitination could participate in the
amino acid signal in mTORC1 sensing microenvironment through RagA [9, 10]. In 2017, Professor Wei Wenyi from Harvard
Medical School found that ubiquitination could participate in the synergistic
regulation of mTORC1 and mTORC2 signaling pathways through GβL [11]. In addition, in 2018, Liu Ying research group
of Institute of molecular medicine of Peking University / Peking University and
Tsinghua University Joint Center for life sciences, found that ubiquitination
participated in the amino acid signal of mTORC1 sensing microenvironment
through the degradation of DEPDC5 (Nature: Liu Ying group made important
progress in mTORC1, tumorigenesis and senescence regulation - Comments from
Professor Liu Wei) [12].
Recently, Wang Ping's research group published a
research paper entitled Ubiquitination of Rheb governs growth factor-induced
mTORC1 activation in Cell Research. This paper expounded that ubiquitination
could participate in mTORC1 sensing growth factor signal in microenvironment
through Rheb.

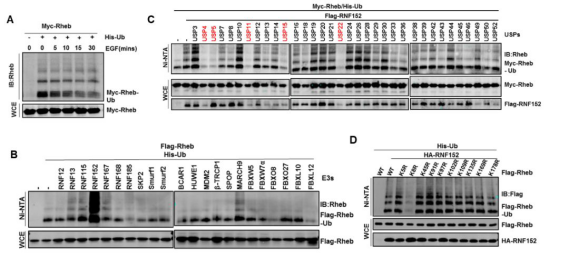
Rheb, as a direct activator of mTORC1, plays a key
role in the activation of mTORC1 [13]. This study found that Rheb not only
could be modified by ubiquitination, but also its ubiquitination was regulated
by growth factor signal, its E3 ubiquitin ligase RNF152, its depubiquitinase
USP4 and its ubiquitin site K8 were identified by screening (as shown in the
figure below).
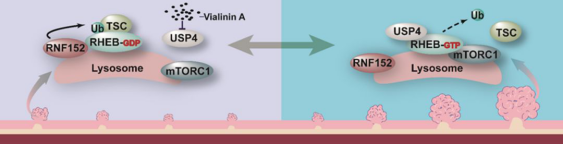
As a post-translational modification, ubiquitination
could regulate the stability, activity or localization of the substrate. This
study found that ubiquitination of Rheb would not affect the stability of Rheb
protein. By detecting the activity of RNF152 and USP4 in MEF, it was found that
ubiquitination could inhibit the activity of small G protease of Rheb. Further
study showed that ubiquitination could inhibit the small G protease activity of
Rheb by promoting the integration of Rheb and TSC2.
At the same time, through the MEF cells of RNF152, USP4
and K8R knockin cell lines, it was found that ubiquitination of Rheb could
regulate the activity, cell proliferation, cell size and autophagy of mTORC1.
In addition, the study also found that ubiquitination of Rheb played an
important role in the occurrence and development of tumors through subcutaneous
tumorigenesis in nude mice and AOM-DSS-induced colorectal cancer model.
In conclusion, this work revealed that ubiquitination
could participate in the growth factor signal in mTORC1 sensing
microenvironment through Rheb, and expounded that ubiquitination of Rheb was an
important regulatory mechanism for the occurrence and development of tumors(as
shown in the figure below).
It was reported that Deng Lu and Chen Lei from Prof. Wang
Ping's research group were the co-first authors of this paper, and Prof. Wang
Ping was the corresponding author.
Reference:
1. Saxton, R.A.
and D.M. Sabatini, mTOR Signaling in Growth, Metabolism, and Disease. Cell,
2017. 169(2): p. 361-371.
2. Dibble, C.C.
and B.D. Manning, Signal integration by mTORC1 coordinates nutrient input with
biosynthetic output. Nat Cell Biol, 2013. 15(6): p. 555-64.
3. Laplante, M.
and D.M. Sabatini, mTOR signaling in growth control and disease. Cell, 2012.
149(2): p. 274-93.
4. Guertin, D.A.
and D.M. Sabatini, Defining the role of mTOR in cancer. Cancer Cell, 2007.
12(1): p. 9-22.
5. Dibble, C.C.
and L.C. Cantley, Regulation of mTORC1 by PI3K signaling. Trends Cell Biol,
2015. 25(9): p. 545-55.
6. Komander, D.
and M. Rape, The ubiquitin code. Annu Rev Biochem, 2012. 81: p. 203-29.
7. Rajalingam, K.
and I. Dikic, SnapShot: Expanding the Ubiquitin Code. Cell, 2016. 164(5): p.
1074-1074 e1.
8. Heride, C., S.
Urbe, and M.J. Clague, Ubiquitin code assembly and disassembly. Curr Biol,
2014. 24(6): p. R215-20.
9. Deng, L., et
al., The ubiquitination of rag A GTPase by RNF152 negatively regulates mTORC1
activation.Mol Cell, 2015. 58(5): p. 804-18.
10. Jin, G., et
al., Skp2-Mediated RagA Ubiquitination Elicits a Negative Feedback to Prevent
Amino-Acid-Dependent mTORC1 Hyperactivation by Recruiting GATOR1. Mol Cell,
2015. 58(6): p. 989-1000.
11. Wang, B., et
al., TRAF2 and OTUD7B govern a ubiquitin-dependent switch that regulates mTORC2
signalling. Nature, 2017. 545(7654): p. 365-369.
12. Chen, J., et
al., KLHL22 activates amino-acid-dependent mTORC1 signalling to promote
tumorigenesis and ageing. Nature, 2018. 557(7706): p. 585-589.
13. Aspuria, P.J.
and F. Tamanoi, The Rheb family of GTP-binding proteins. Cell Signal, 2004.
16(10): p. 1105-12.
